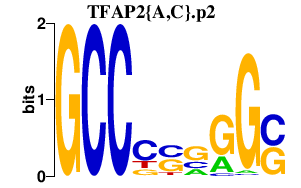
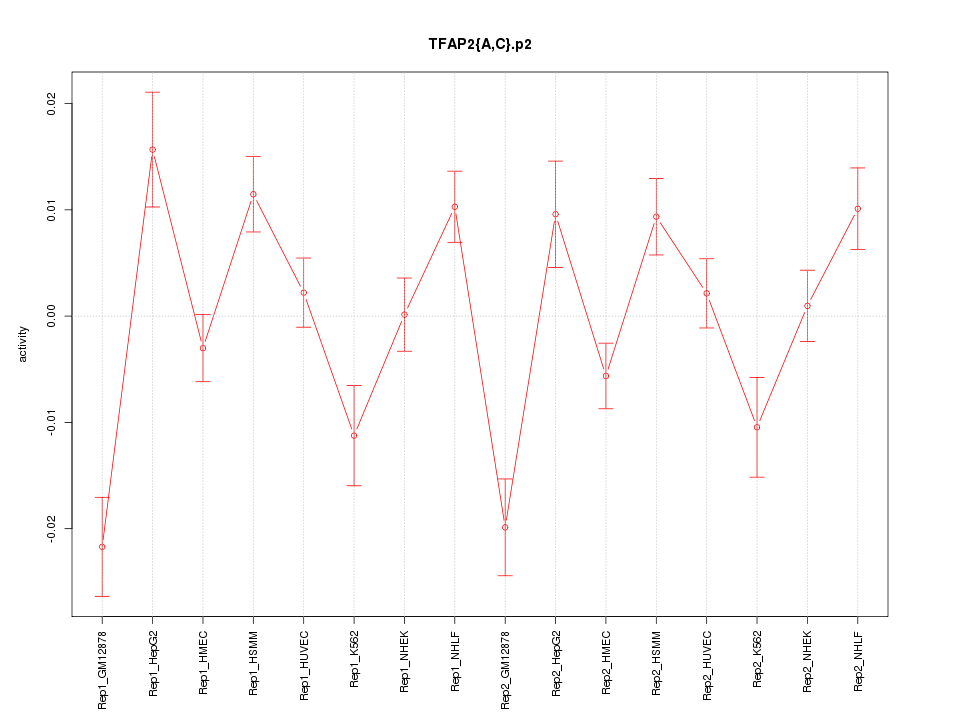
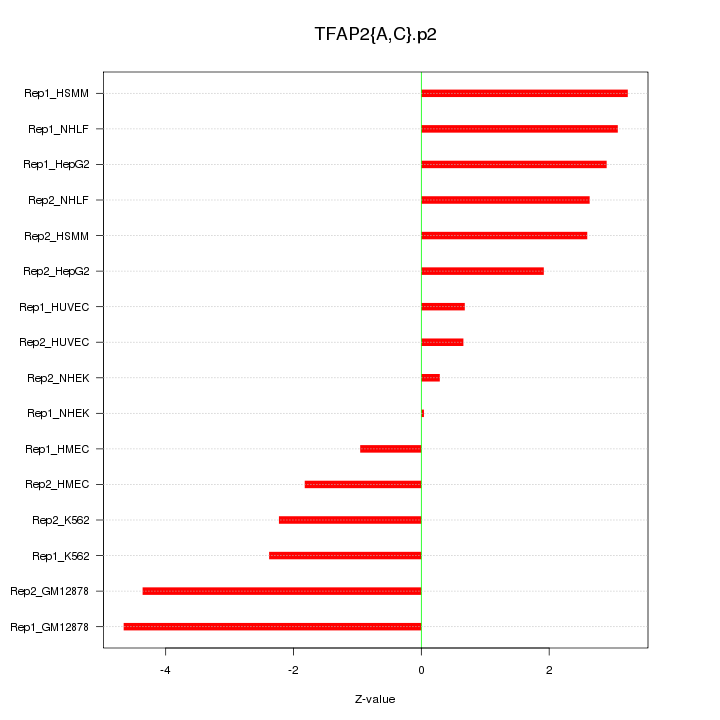
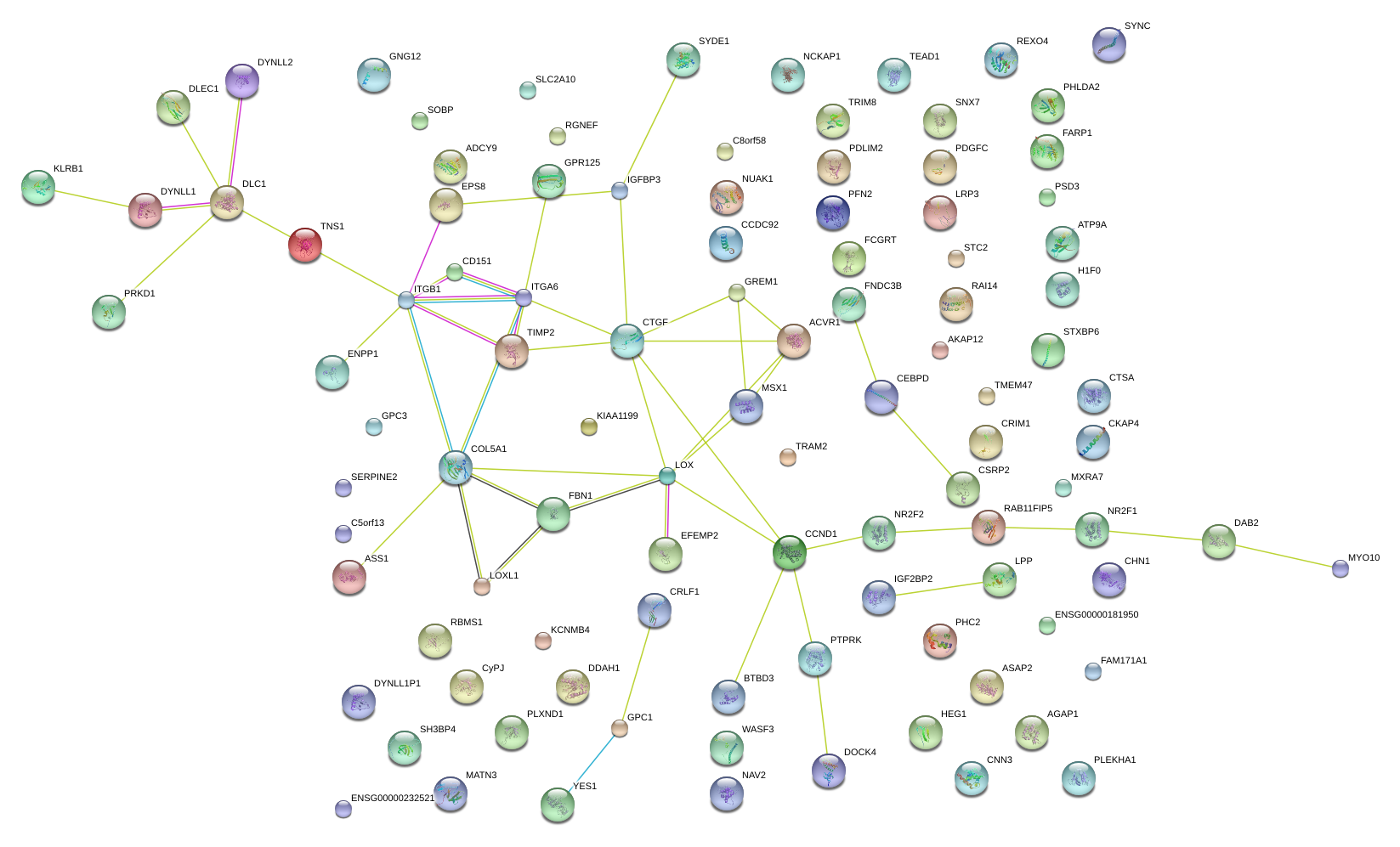
|
chr17_-_72218661
|
7.061
|
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr3_-_187025501
|
6.416
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr17_-_72218535
|
5.021
|
NM_001008528
NM_001008529
NM_198530
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr19_-_18578467
|
4.672
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr19_-_18578631
|
4.557
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr9_+_136673464
|
4.504
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr3_-_187025438
|
4.407
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr2_+_36436873
|
4.160
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr11_-_65396775
|
4.070
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr19_-_18578577
|
4.044
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr15_-_46724360
|
3.846
|
|
FBN1
|
fibrillin 1
|
|
chr1_-_85703198
|
3.784
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_-_224612177
|
3.715
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr3_-_151171190
|
3.618
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr4_-_158111995
|
3.482
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr2_+_235525355
|
3.452
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr7_-_45927263
|
3.447
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr3_-_126257425
|
3.399
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr10_+_104394185
|
3.353
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr3_-_151171576
|
3.347
|
|
PFN2
|
profilin 2
|
|
chr22_-_34754343
|
3.344
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr6_+_151603346
|
3.318
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr12_-_105165805
|
3.278
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr5_+_34692124
|
3.173
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr8_+_22492586
|
3.142
|
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr11_+_12652427
|
3.135
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr3_-_130807865
|
3.125
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr5_+_92946348
|
3.124
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr6_+_107917965
|
3.014
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr2_+_173000590
|
2.979
|
|
ITGA6
|
integrin, alpha 6
|
|
chr12_-_123023312
|
2.912
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr6_-_128883125
|
2.908
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr20_+_44771532
|
2.893
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr10_+_124124196
|
2.881
|
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr5_+_34692352
|
2.764
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr9_+_132309914
|
2.748
|
NM_000050
NM_054012
|
ASS1
|
argininosuccinate synthase 1
|
|
chr22_+_36531246
|
2.726
|
|
H1F0
|
H1 histone family, member 0
|
|
chr12_-_15833617
|
2.674
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr13_+_26029799
|
2.632
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr5_+_34692239
|
2.611
|
|
RAI14
|
retinoic acid induced 14
|
|
chr8_-_48813256
|
2.605
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr6_+_151603201
|
2.584
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr2_+_236067424
|
2.551
|
NM_001037131
NM_014914
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr2_-_218551786
|
2.533
|
|
TNS1
|
tensin 1
|
|
chr8_-_48813236
|
2.521
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr13_+_97593711
|
2.515
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr6_-_52549774
|
2.499
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr2_+_173000552
|
2.473
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr11_-_2907169
|
2.436
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chrX_-_34585287
|
2.432
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr6_-_132314004
|
2.416
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr2_-_20075889
|
2.385
|
|
MATN3
|
matrilin 3
|
|
chr5_-_16989371
|
2.373
|
NM_012334
|
MYO10
|
myosin X
|
|
chr1_-_95165037
|
2.364
|
|
CNN3
|
calponin 3, acidic
|
|
chr19_+_15079141
|
2.291
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr2_-_158440587
|
2.290
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr12_+_69046241
|
2.270
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr20_+_11819364
|
2.260
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr13_+_26029879
|
2.256
|
|
WASF3
|
WAS protein family, member 3
|
|
chr12_-_75796898
|
2.251
|
NM_001321
|
CSRP2
|
cysteine and glycine-rich protein 2
|
|
chr5_-_172688024
|
2.251
|
|
STC2
|
stanniocalcin 2
|
|
chr7_-_111633620
|
2.246
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr1_-_32940943
|
2.226
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr2_-_183610992
|
2.190
|
|
NCKAP1
|
NCK-associated protein 1
|
|
chr4_-_22126437
|
2.180
|
NM_145290
|
GPR125
|
G protein-coupled receptor 125
|
|
chr15_+_94674849
|
2.144
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr17_-_74432800
|
2.141
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr2_-_160972606
|
2.133
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr10_-_15453060
|
2.090
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr1_-_85703321
|
2.090
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr14_-_29466562
|
2.087
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr6_-_52549673
|
2.071
|
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr16_-_4106022
|
2.071
|
|
ADCY9
|
adenylate cyclase 9
|
|
chr15_+_72006015
|
2.058
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr3_+_173240983
|
2.056
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr12_-_105056577
|
2.051
|
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chrX_-_132947141
|
2.048
|
|
GPC3
|
glypican 3
|
|
chr12_-_15833383
|
2.025
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr8_-_18915443
|
2.006
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr1_-_33587998
|
1.996
|
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr11_+_69165018
|
1.973
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr2_+_241023703
|
1.954
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr1_-_68071635
|
1.950
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr10_-_33287137
|
1.938
|
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr15_+_72005875
|
1.932
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr11_+_822965
|
1.897
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr16_-_4106186
|
1.895
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr15_+_78858763
|
1.892
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chr19_+_38377329
|
1.890
|
NM_002333
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr2_-_73193566
|
1.879
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr3_-_45242072
|
1.872
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr5_-_121441910
|
1.867
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr8_-_13035102
|
1.859
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr14_-_24588916
|
1.841
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr4_+_4912272
|
1.835
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr5_-_111121691
|
1.825
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_98899734
|
1.814
|
NM_015976
NM_152238
|
SNX7
|
sorting nexin 7
|
|
chr2_-_73193419
|
1.809
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr3_+_189354167
|
1.806
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr2_+_9264360
|
1.796
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr5_-_39460687
|
1.785
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr6_+_132170888
|
1.771
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr15_+_30797466
|
1.769
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr20_-_49818268
|
1.762
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr2_-_175578156
|
1.755
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr22_+_36531059
|
1.750
|
NM_005318
|
H1F0
|
H1 histone family, member 0
|
|
chr2_-_73193581
|
1.746
|
NM_015470
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr18_-_802268
|
1.745
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr1_-_33587927
|
1.738
|
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr10_+_124124083
|
1.735
|
NM_001001974
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chrX_-_132947290
|
1.725
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr19_+_54708247
|
1.718
|
NM_004107
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr20_+_43952997
|
1.717
|
NM_000308
NM_001127695
NM_001167594
|
CTSA
|
cathepsin A
|
|
chr11_+_19691397
|
1.713
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr5_+_72957736
|
1.707
|
NM_001080479
NM_001177693
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr6_+_148705646
|
1.685
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr11_+_129445002
|
1.684
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr5_-_57791544
|
1.679
|
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr18_-_24011349
|
1.676
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr2_+_101680594
|
1.662
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr5_-_39460787
|
1.651
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr5_-_39461054
|
1.643
|
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr4_-_57671112
|
1.641
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr10_-_33287184
|
1.630
|
NM_002211
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr1_-_33588085
|
1.624
|
NM_004427
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr22_+_36531178
|
1.621
|
|
H1F0
|
H1 histone family, member 0
|
|
chr11_+_129445017
|
1.621
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr22_+_31527744
|
1.597
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr3_+_37878647
|
1.590
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr3_-_121652183
|
1.588
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr6_+_1336077
|
1.564
|
|
|
|
|
chr10_+_29006425
|
1.558
|
NM_012342
|
BAMBI
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis)
|
|
chr8_+_37773553
|
1.553
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr20_-_60375695
|
1.548
|
NM_005560
|
LAMA5
|
laminin, alpha 5
|
|
chr2_-_55130963
|
1.546
|
|
RTN4
|
reticulon 4
|
|
chr6_-_138470160
|
1.536
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr10_+_123912930
|
1.534
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr8_+_30361485
|
1.533
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chrX_+_117745551
|
1.527
|
NM_001560
|
IL13RA1
|
interleukin 13 receptor, alpha 1
|
|
chr13_-_109757442
|
1.510
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr5_-_121441866
|
1.510
|
|
LOX
|
lysyl oxidase
|
|
chr8_+_37773564
|
1.485
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr2_+_46377952
|
1.483
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr14_-_74148422
|
1.478
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr2_-_20075929
|
1.470
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr10_-_79356285
|
1.462
|
NM_004747
|
DLG5
|
discs, large homolog 5 (Drosophila)
|
|
chr2_-_45090025
|
1.462
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr10_+_134201262
|
1.456
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr16_-_51138306
|
1.453
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr2_-_220116508
|
1.450
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr17_-_74433048
|
1.440
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr1_-_94779727
|
1.429
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr2_-_55130925
|
1.424
|
|
RTN4
|
reticulon 4
|
|
chr2_+_113119985
|
1.415
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr1_+_2149993
|
1.411
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr1_+_111217244
|
1.406
|
NM_000560
|
CD53
|
CD53 molecule
|
|
chr4_-_1156458
|
1.404
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr17_-_60088459
|
1.398
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr13_-_44048601
|
1.398
|
NM_183422
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr15_+_72005864
|
1.398
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr17_+_72376022
|
1.395
|
NM_144677
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr16_-_88084387
|
1.395
|
NM_013275
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chr7_-_107430566
|
1.393
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr1_+_111217314
|
1.391
|
|
CD53
|
CD53 molecule
|
|
chr11_+_822909
|
1.389
|
NM_001039490
NM_004357
NM_139029
NM_139030
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr20_+_29656744
|
1.388
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr9_-_83493415
|
1.386
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr13_+_109757593
|
1.382
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr3_-_38666122
|
1.378
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr2_+_96845704
|
1.371
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr4_-_1156378
|
1.366
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr11_+_129444997
|
1.364
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr4_-_1156533
|
1.359
|
NM_012445
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr22_+_31527678
|
1.357
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr4_+_38545741
|
1.353
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr20_-_49818306
|
1.351
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr17_-_77411607
|
1.351
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr15_-_88159061
|
1.343
|
NM_001150
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr11_-_63292688
|
1.337
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr1_-_1157284
|
1.336
|
NM_016176
NM_016547
|
SDF4
|
stromal cell derived factor 4
|
|
chr9_+_123070234
|
1.335
|
|
GSN
|
gelsolin
|
|
chr1_+_19843261
|
1.330
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr19_+_40213431
|
1.329
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr3_+_180853473
|
1.327
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr1_-_120413542
|
1.308
|
|
NOTCH2
|
notch 2
|
|
chr6_+_132170847
|
1.307
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr15_+_65145248
|
1.305
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr22_-_27405708
|
1.303
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr1_-_30977931
|
1.301
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr19_+_50004134
|
1.299
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr6_+_146906520
|
1.283
|
NM_006834
|
RAB32
|
RAB32, member RAS oncogene family
|
|
chr2_-_224611568
|
1.282
|
NM_001136530
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr2_-_101134146
|
1.281
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr1_-_120413798
|
1.281
|
NM_024408
|
NOTCH2
|
notch 2
|
|
chr3_+_51403760
|
1.279
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr20_-_47238310
|
1.277
|
NM_001037328
NM_004602
NM_017452
NM_017453
NM_017454
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr21_+_17807167
|
1.265
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr8_-_62789665
|
1.260
|
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr3_-_45242624
|
1.255
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr11_+_116575249
|
1.249
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|